Abstract
Background
In developed nations, myocardial infarction (MI) is one of the main causes of morbidity and mortality, resulting in a significant economic burden and becoming a global public health problem. C1q/tumor necrosis factor-related protein 9 (CTRP9) is a secreted protein comprising a variable domain, a collagenous region, and a C-terminal trimerizing globular C1q (gC1q) domain. In vivo, the full-length CTRP9 (fCTRP9) can be cleaved into the globular domain of CTRP9 (gCTRP9). Here, we tested the cardio-protective impacts of fCTRP9, gCTRP9, and N-terminal domain, including the variable and collagenous domain, of CTRP9 (nCTRP9) in the context of MI.
Methods
Studies comparing the protective properties of fCTRP9 and gCTRP9 against MI in mice hearts were performed both in vitro and in vivo. The role of matrix metalloproteinase-9 (MMP9) in CTRP9 cleavage was examined, and the effects of different CTRP9 domains on cardiac fibrosis and cardiac fibroblast (CF) activation were investigated.
Results
gCTRP9 exerted better protective effects than fCTRP9 against MI, demonstrating superior anti-apoptotic and anti-fibrotic properties. fCTRP9 was cleaved by MMP9, resulting in gCTRP9 and nCTRP9. MMP9 overexpression enhanced the cardioprotective effects of fCTRP9, while nCTRP9 supplementation aggravated cardiac fibrosis in MI mice. Mechanistically, nCTRP9 activated CFs via an increase in Rap1 expression and MEK 1/2 and ERK1/2 phosphorylation.
Conclusions
Different domains of CTRP9 have distinct cardioprotective effects. gCTRP9 shows beneficial effects, while nCTRP9 promotes cardiac fibrosis. These findings highlight the importance of CTRP9 in cardiac function regulation and suggest prospective therapeutic options for MI treatment.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Myocardial infarction (MI) is the most severe clinical manifestation of coronary artery disease and a potentially fatal coronary event that results in rapid cardiac death. It is the primary cause of heart failure globally and a significant cause of morbidity and mortality in developed nations. Consequently, it has a substantial financial impact and is becoming a global public health concern [1, 2].
Adipose tissue functions as an endocrine organ by secreting proeins known as adipokines, such as leptin, resistin, retinol-binding protein4, angiopoietin-like protein2, interleukin-6, adiponectin and c1q/tumor necrosis factor-related proteins (CTRPs) [3]. These adipokines play either beneficial or adverse roles in cardioprotection after MI. The CTRPs family consists of 15 members, CTRP1-CTRP15. CTRP9 was the first reported to have a protective effect on ischemic myocardium [4]. After MI, the CTRP9 expression in the myocardium is significantly decreased, whereas exogenous supplementation with CTRP9 can play a protective role by alleviating pathological myocardial remodeling after MI [4]. Previous research has indicated that CTRP9 necessitates proteolytic cleavage to produce a physiologically active globular domain isoform (gCTRP9) but not full-length CTRP9 (fCTRP9), which can activate important cardiac survival kinases such as Akt, AMPK, and eNOS [5]. The aims of the current study were to determine whether fCTRP9 and its lysate products play the same role in the pathological remodeling process after MI.
Methods
Animals
Eight-week-old male C57BL/6 adult mice were bought from the Fourth Military Medical University’s Experimental Animal Center in Xi’an, China. The Fourth Military Medical University Committee of Animal Care authorized all experimental methods, which complied with the National Institute of Health Guidelines for the Care and Use of Laboratory Animals (No. IACUC-20210612). Each animal had unrestricted access to standard food and water. The mice were kept in a light-controlled environment with a 12-hour light/dark cycle, with 22–24 °C temperature and a 30–70% humidity range. Mice were randomly assigned to differernt group using a random number table. Briefly, mice were sequentially numbered based on their body weight. A set of random numbers (equal to totle number of mice) was then obtained from a random number table, starting from an arbitrary point and proceeding in a single direction. These random numbers were divided by the total number of groups, and mice were assigned to groups according to the remainder. If any group did not reach its pre-determined number, additional mice were allocated using the subsequent random number to ensure that each group met its required sample size. When investigating the effects of different forms of CTRP9 on MI, mice were randomized into the following groups: (1) Sham (n = 10); (2) MI + Saline (n = 15); (3) MI + fCTRP9 (n = 15); and MI + gCTRP9 (n = 15). When investigating the effects of MMP9 over-expression on fCTRP9 against MI, mice were randomized as follows: (1) NC + Saline (n = 15); (2) NC + fCTRP9 (n = 15); (3) MMP9 OE + Saline (n = 15); and (4) MMP9 OE + fCTRP9 (n = 15). When investigating the effects of nCTRP9 against MI, mice were randomized as follows: (1) Sham + Saline (n = 14); (2) Sham + nCTRP9 (n = 14); (3) MI + Saline (n = 20); and (4) MI + nCTRP9 (n = 20). When investigating the effects of Rap1 knock-down on nCTRP9 against MI, mice were randomized as follows: (1) shNC + MI + Saline (n = 20); (2) shNC + MI + nCTRP9 (n = 20); (3) shRap1 + MI + Saline (n = 20); and (4) shRap1 + MI + nCTRP9 (n = 20). Team members were assigned to Group A and Group B based on their skills and experience in accordance with the frameworks of this study. Group A was primarily responsible for randomization, blinding, and ensuring animal welfare, while Group B was tasked with executing the experiment, as well as collecting and analyzing the data. Members from both groups could make minor adjustments before the commencement of each phase of the study.
MI surgery
As per the previously described process, MI was produced by ligating the mice’s left coronary artery [6]. Briefly, 3% isoflurane (R510-22-10, RWD Life Science, Shenzhen, China) was used to induce anesthesia in the mice, followed by 2% isoflurane for the maintenance of anesthesia in an anesthesia induction chamber. Mice were fixed on the surgery table, followed by endotracheal intubation, with the tidal volume (150 µl) and respiratory rate (100 times /min) adjusted. Mouse hearts were exposed from the intercostal space, and a 6 − 0 silk suture was used to ligate the left coronary artery about 3 mm from its origin. The identical treatment was performed on mice in the Sham group without suture ligation. The mice were transferred to an incubator until full recovery from anesthesia after the surgery. Carprofen (5 mg/kg) was administered prior to awakening and and was re-administered 24 h postoperatively. The condition of the mice, including wound status, was closely monitored, and appropriate symptomatic treatment was provided if any abnormalities were observed.
Echocardiography
A high-resolution in vivo imaging system (Vevo 2100, VisualSonics, Toronto, Canada) with an MS-400 imaging transducer was used to perform echocardiography for cardiac assessment as described previously [7, 8]. Briefly, mice were carefully placed on a temperature-controlled pad (42 °C) after being given isoflurane (R510-22-10, RWD Life Science, Shenzhen, China) for anesthesia. A pre-warmed medical ultrasonic couplant was placed on the shaved chest. The probe was then rotated counterclockwise by approximately 15° to obtain a parasternal long-axis view after the probe placed vertically with the notch facing the mouse’s head. The apex of the heart and the outflow tract are at the same level. Then rotate the probe 90° to obtain a short-axis view and recorded for measurement. Short-axis views of the M-mode of the left ventricle were recorded for measurement and calculation using Vevo Lab 3.1.0 software (FUJIFILM VisualSonics, Toronto, Canada). Echocardiographic data were acquired by two independent examiners and measured by two separate individuals. Cardiac parameters were reported as the mean of the measurements obtained from three consecutive cardiac cycles, after excluding any respiratory influence.
Expression and purification of CTRP9
Variants of human CTRP9, fCTRP9, gCTRP9, and N-terminal domain, including the variable and collagenous domain, of CTRP9 (nCTRP9) were expressed in Escherichia coli and purified as previously described [9]. Gene synthesis was performed according to E. coli codon preference optimization, using the appropriate region of C1QTNF9 (GenBank Accession No. NM_178540.5), 143 bp to 1087 bp for fCTRP9, 143 bp to 670 bp for nCTRP9, and 671 bp to 1087 bp for gCTRP9. The synthetic DNA fragment was cloned into the expression vector pET30a followed by transformation into E. coli BL21(DE3). BL21(DE3) with recombinant plasmid was grown in Luria-Bertani liquid medium for protein expression via the addition of isopropyl-β-D-thiogalactoside to a final concentration of 1 mM at 16 °C for 24 h. A nickel-affinity chromatography column (17524801, Cytia, Massachusetts, USA) was used for CTRP9 purification from the lysate of BL21(DE3) with recombinant plasmid, in compliance with the manufacturer’s guidelines. Using an endotoxin removal column (88276; Thermo Fisher Scientific, Carlsbad, CA, USA), the endotoxins were removed. Purity, as evaluated by SDS-PAGE with Coomassie Brilliant Blue staining, was confirmed to be greater than 90%, which is considered sufficient for use. The yield of fCTRP9 and nCTRP9 exceeded 2 mg per liter of E. coli culture medium, whereas the yield of gCTRP9 was approximately 500 µg per liter of E. coli culture medium.
fCTRP9 digestion by matrix metalloproteinase-9 (MMP9) in vitro
The procedure for fCTRP9 digestion by MMP9 was done as previously mentioned [10]. The digestion system consisted of 4 µg fCTRP9 protein, 1 µg MMP9 (CI71, Novoprotein, SuZhou, China), 10 µL reaction buffer comprising 200 mM Tris HCl (pH8.0), 1500 mM NaCl (S8210, Solarbio, Beijing, China), 50 mM CaCl2 (10035-0, Sigma-Aldrich, St. Louis, MO, USA), and 1 mM ZnCl2 (Z298947, Aladdin Biochemical Technology, Shanghai, China). All reagents were mixed thoroughly and incubated at 37℃ for 40 min; products were detected by SDS-PAGE.
Histological analysis
Isoflurane anesthetized mice blood was collected from the carotid artery and then their hearts were removed immediately. The hearts were fixed in paraformaldehyde (4%; G1101, Servicebio, Wuhan, China) at 4 °C for about 24 h for histological analysis. Following the prior procedure, the heart tissue was embedded in paraffin and sliced into 6 μm thick sections from the cardiac apex for staining with either wheat germ agglutinin (WGA; L4895, Sigma-Aldrich, St. Louis, MO, USA) or Masson (CD069, ZHHC, Xi’an, China) [8]. ImageJ software (National Institutes of Health) was used to measure the infarct size and fibrosis using Masson’s trichrome staining. Infarct size was determined as the percentage of midline infarct lengths occupied by midline circumferences from heart section. WGA-stained images were captured using an inverted confocal microscope (Carl Zeiss, Jena, Germany), and using ImageJ, the surface area of cardiomyocytes was determined.
Neonatal rat cardiomyocyte (NRCM) isolation and culture
As previously mentioned, NRCMs were isolated using an enzyme process from the ventricles of newborn (1–3 days) Sprague Dawley (SD) rats [11]. Briefly, tissue fragments of the left ventricle were digested with 0.1% trypsin (1004GR025; BioFroxx, Einhausen, Germany) and 1% collagenase II (2275GR001; BioFroxx, Einhausen, Germany). Subsequently, the resuspended NRCMs were inoculated in plates or dishes after differential adhesion in DMEM/F12 containing 0.1 mM bromodeoxyuridine for 1.5–2 h. NRCMs were inoculated in 96-well plates or dishes, treated with 15 ng/mL fCTRP9, nCTRP9, or gCTRP9 for 18 h, and placed in a Napco 8000WJ hypoxia incubator (Thermo Fisher Scientific, Carlsbad, CA, USA) for 6 h. The Cell Counting Kit-8 (CCK-8; C0005, Topscience, Boston, MA, USA) assay was used to evaluate the cell viability of NRCMs in accordance with the manufacturer’s instructions. After fixing in 4% paraformaldehyde for 10 min, TUNEL staining was conducted using an in situ cell death detection kit (11684817910; Roche, Basel, Switzerland) per the directions provided by the manufacturer.
Generation of immortalized cardiac fibroblasts (iCF)
iCFs were generated as previously described [12]. Mouse hearts were isolated and rinsed with ice-cold HBSS (H1025, Solarbio, Beijing, China). The tissue fragments were successively digested with 0.4% trypsin and 1% collagenase II (2275GR001; BioFroxx, Einhausen, Germany). Cardiac fibroblasts (CFs) were infected with 293T supernatant containing lentivirus, followed by selection with puromycin (10 µg/mL). The supernatant containing the lentivirus was generated using the SV40 T Antigen Cell Immortalization Kit (CILV01, Alstem, San Francisco, USA) according to the manufacturer’s guidelines. Following the manufacturer’s suggested directions, three plasmids (psPAX2, SV40 LT, and PMD2.G) were co-transfected into 293T cells using Lipofectamine LTX and Plus Regent (15338-100, Thermo Fischer Scientific, Carlsbad, CA, USA). After 72 h of culture, the supernatant containing the lentivirus was collected and used to generate iCFs.
Following the instructions given by the manufacturer, the BeyoClick™ EdU Cell Proliferation Kit (C0078S, Beyotime, Shanghai, China) was used to perform ethynyl-2’-deoxyuridine (Edu) staining, which measured cell proliferation. After the addition of the EdU reagent to the medium for 2 h, the sample was fixed for 10 min in paraformaldehyde (4%) with Triton X-100 (0.1%). Subsequently, 250 µL/well clicking solution was added and incubated for 30 min. DAPI (62248, Thermo Fisher Scientific, Carlsbad, CA, USA) was then used to stain the nuclei. Using a Zeiss inverted confocal microscope, the fluorescence of the EdU staining was captured, and ImageJ software was used to evaluate the results.
Cell viability assay
Cell growth assays were performed using a Cell Counting Kit-8 (CCK-8; Beyotime Institute of Biotechnology, Nanjing, China) based on the manufacturer’s protocol. Cells.
were incubated with 10% CCK-8 solution in 96-well plates for 2–4 h at 37 °C. The absorbance was detected at 450 nm using a microplate spectrophotometer.
Cell proliferation assay
The 5-ethynyl-2-deoxyuridine (EdU)incorporation assay was carried out using an EDU assay kit (BeyoClick™ EdU-555, Beyotime, C0075S) following the manufacturer’s instructions. Images were photographed with a Zeiss LSM 900 microscope (Carl Zeiss, Germany). iCF proliferation was quantified as the percentage of EdU-positive cells.
SiRNA transfection
siRNA for repressor activator protein 1 (Rap1) was provided by Tsingke Biotechnology Co., Ltd. (Beijing, China). Following the manufacturer’s recommendations, 50 µM siRNA was administered to iCFs that had reached about 75% confluence. The Lipofectamine™ Reagent (MAN0014545, Thermo Fischer Scientific, Carlsbad, CA, USA) was used for this process. Following a 48 h siRNA incubation, the iCFs were subjected to Tgf-β (10 ng/mL, 100 − 21, Proteintech, Wuhan, China), fCTRP9 (15 ng/mL), gCTRP9 (15 ng/mL), or nCTRP9 (15 ng/mL) for 24 h. Following this, the cells were collected and prepared for further examination.
RNA-seq
Total RNA was extracted from iCF cells using an RNAprep Pure Kit (DP432; TIANGEN Biotech, Beijing, China) following the procedure provided by the manufacturer. RNA-seq and bioinformatic analyses were performed by Standard Sci-Tech Innovation (Qingdao) Pharmaceutical Technology Co., Ltd. (Qingdao, China) [13]. The RNA libraries were constructed with the VAHTS mRNA-seq V3 Library Prep Kit (NR604, Vazyme, Nanjing, China) for Illumina sequencing using 1 µg of total RNA. Afterward, the Qsep1 equipment (Bioptic, Changzhou, China) was employed to evaluate the integrity of RNA samples. The raw data underwent routine bioinformatics analysis with a filter threshold of fold change (FC) > 1.2.
Western blot
Western blotting was done in accordance with earlier instructions [14]. Using RIPA lysis buffer (R0010; Solarbio, Beijing, China), the whole protein was extracted. Following the instructions provided by the manufacturer, the total protein concentration was measured using a BCA total protein quantification kit (P0009, Beytome, Shanghai, China). Using a pre-stained protein ladder (SW175-04; Sevenbio, Beijing, China) as an indicator, total proteins were separated by gel electrophoresis and then transferred onto PVDF membranes. Subsequently, the membranes were treated for 1 h at room temperature with the relevant secondary antibodies after overnight incubation at 4 °C with the suitable primary antibodies. The following are the primary antibodies: CTRP9 (1:1000, customized by Genscript), nCTRP9 (1:1000, customized by Genscript), Rap1 (1:1000, 68125-1-Ig, Proteintech, Wuhan, China) GAPDH (1:5000, AC001, Abclonal, Wuhan, China), AMPK (1:1000, 2532, CST, Danvers, USA) and p-AMPKαThr172 (1:1000, 2532, CST, Danvers, USA), Phospho-p38 MAPKαThr 172 (1:1000, 4060, CST, Danvers, USA), Akt (1:1000, 9272, CST, Danvers, USA)and p-Akt Ser 473 (1:1000, 4060, CST, Danvers, USA), ERK1/2 (1:1000, 4696, CST, Danvers, USA), p-ERK1/2 Thr202/Tyr204 (1:1000, 9106, CST, Danvers, USA), MEK1/2 (1:1000, 11049-1-AP, Proteintech, Wuhan, China)and Phospho-MEK1/2(1:1000, 9154, CST, Danvers, USA). Secondary antibodies used were rabbit IgG (ZB-2301; ZSGB-BIO, Beijing, China) and mouse IgG (ZB-2305; ZSGB-BIO). A Bio-Rad imaging system (Hercules, California, USA) was used for observing the blots using ECL-plus reagent, and Image Lab 5.1 software (Bio-Rad Laboratories, Hercules, CA, USA) was utilized for analysis.
RNA isolation and quantitative PCR
Quantitative PCR (qPCR) and RNA isolation were carried out according to a previously published protocol [15]. Briefly, total RNA was extracted via a total RNA extraction kit (DP419; TIANGEN Biotech, Beijing, China). After detecting RNA concentration, 1 µg RNA was used for reverse transcription with StarLighter Script RT all-in-one Mix (FS-P1002, Forever star, Beijing, China). StarLighter SYBR Green qPCR Mix (FS-Q1002; Forever Star; Beijing, China) was used in the qPCR, which was carried out using a CFX96 real-time PCR equipment (C1000 Thermal Cycler). Using the 2−ΔΔCt method, mRNA transcription was calculated and normalized to Actb. Table S1 contains a list of primer sequences used in this investigation.
Adeno-Associated virus (AAV) infection
AAV9 encoding MMP9 or Rap1 shRNA was provided by Tsingke Biotechnology Co., Ltd. (Beijing, China) and is listed in Table S2. Mice received a single injection of recombinant AAV9 or AAV encoding NC at a dosage of 1 × 1011 viral genomes through the tail vein two weeks before MI surgery.
Statistical analysis
Unless otherwise specified, each experiment was repeated at least three times. The data was shown as the mean ± standard deviation (Mean ± SD). Statistical analyses were performed using GraphPad Prism 9.0. (GraphPad Software Inc., La Jolla, USA). When determining the significance of differences, p < 0.05 was deemed statistically significant. It was assessed using an unpaired, two-tailed Student’s t-test for two groups or a one-way ANOVA with Tukey’s multiple comparison test for three or more groups, followed by Bonferroni post hoc analysis.
Results
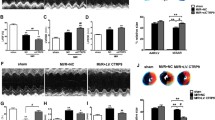
gCTRP9 exerts better protective effect than fCTRP9 on mice heart against MI
CTRP9 comprises four distinct domains: signal peptide, variable region, collagenous domain, and globular domain in C-terminal, whereas mature full-length CTRP9 contains no signal peptides (Fig. 1A). To assess whether fCTRP9 and gCTRP9 exert different effects on infarcted hearts, either fCTRP9 or gCTRP9 was administered following MI surgery for a duration of four weeks (Fig. 1A). The administration of gCTRP9 resulted in significant improvements in cardiac systolic function, as evidenced by an increase in ejection fraction (EF) and a decrease in left ventricular end-diastolic diameter (LVEDD) (Fig. 1B-C). While fCTRP9 also enhanced EF and reduced LVEDD, gCTRP9 demonstrated superior cardioprotective effects compared to fCTRP9 regarding the improvement of EF (Fig. 1B-C). Both fCTRP9 and gCTRP9 treatment led to reductions in the lung weight/body weight ratio (LW/BW) following MI surgery (Fig. 1D). Masson staining revealed that both forms of CTRP significantly decreased infarct size in MI mice heart (Fig. 1E). Furthermore, both treatments mitigated cardiac hypertrophy induced by MI; this was indicated by decreases in heart weight/tibial length ratio (HW/TL) and mean cross-sectional area (CSA) measurements (Fig. 1F-G). Additionally, both fCTRP9 and gCTRP9 were effective in reducing cardiac fibrosis resulting from MI surgery as shown through histological analysis (Fig. 1H-J). Although both gCTRP9 and fCTRP9 exhibited cardioprotective properties, it is noteworthy that gCTRP9 resulted in lower values for infarct size, HW/TL ratio, CSA measurements, and levels of cardiac fibrosis when compared to those observed with fCTRP9 treatment(Figs. 1E-J). These findings suggest that while both forms can confer protection against myocardial injury, gCTRP9 demonstrates superior efficacy. Moreover, administration of fCTRP9 led to an increase in serum levels of gCTRP9 while administration of gCTRP9 did not alter serum levels of fCTRP9 among treated mice (Figure K ). The increased levels of gCTRP9 after fCTRP9 administration imply that fCTRP9 may undergo cleavage into its active form, gCTRP9, in vivo.
gCTRP9 exerts better protective effect than fCTRP9 on mice heart against MI. A. The structure of CTRP9 protein and fCTRP9 or gCTRP9 was administered after MI surgery for 4 weeks. B-C. Impact of different type of CTRP9 on cardiac ejection fraction (EF) (B) and left ventricular end diastolic diameter (LVEDD) (C) of mice post MI surgery. n = 6. D. Lung weight (LW) to body weight (BW) ratio. n = 6. E. Infarct area of mice heart tissue. n = 6. F. Heart weight (HW) to tibial length (TL) ratio. n = 6. G. Staining with wheat germ agglutinin was done to determine the mean cross-sectional area of cardiomyocytes. n = 6. H. Cardiac fibrosis in Sham and MI mice heart detected by Masson staining of the border region and distant region. n = 6. K. fCTRP9 and gCTRP9 level in mice serum. n = 4. * p < 0.05, ** p < 0.01
gCTRP9 exerts anti-fibrotic effect, while fCTRP9 promotes fibrosis in vitro
To investigate the differential cardioprotective effects of gCTRP9 and fCTRP9, we assessed their anti-apoptotic properties. Both gCTRP9 and fCTRP9 enhanced cell viability and reduced apoptosis in simulated ischemic (SI) NRCMs; however, gCTRP9 exhibited more pronounced protective effects (Fig. 2A-C). Additionally, we evaluated the anti-fibrotic effects of both proteins. iCFs were generated to assess the impact of gCTRP9 and fCTRP9 on fibrosis (Fig. 2D). Treatment with gCTRP9 significantly diminished Tgf-β-induced proliferation of iCFs s as measured by EdU staining (Fig. 2E). Furthermore, gCTRP9 attenuated CFs activation, indicated by a reduction in α-SMA expression and decreased transcription levels of fibrosis-related genes (Figures F-H). In contrast, fCTRP9 did not inhibit the proliferation or activation of iCFs (Fig. 2I-M). Surprisingly, treatment with fCTRP9 exacerbated Tgf-β-induced proliferation and activation of ICFs. These findings suggest that while gCTRP9 exhibits an anti-fibrotic effect, fCTRP9 appears to promote fibrogenesis in vitro.
gCTRP9 exerts anti-fibrotic effect while fCTRP9 promotes fibrosis on Cardiac fibroblasts. A. gCTRP9 or fCTRP9 was administrated to primary cardiomyocytes after simulated ischemic (SI) treatment. B-C. Both gCTRP9 and fCTRP9 exerts protective effect on SI cardiomyocytes via increasing cell viability (B) and decreasing apoptosis (C). n = 6. D. Mice cardiac fibroblasts (CF) were first induced to immortalized cardiac fibroblasts (iCF), followed by activated with Tgf-β, and finally effect of fCTRP9 on fibrosis was tested. E-H. gCTRP9 inhibits the activation of iCF by reducing cell proliferation (E), n = 6, decreasing α-smooth muscle actin (α-SMA) expression (F-G), n = 6, and decreasing transcription of fibrosis-related genes (H), n = 4. I-K. fCTRP9 promotes the activation of iCF by promoting cell proliferation (J-K), n = 6, increasing α-smooth muscle actin (α-SMA) expression (L), n = 6, and enhancing transcription of fibrosis-related genes (M), n = 4. * p < 0.05, ** p < 0.01
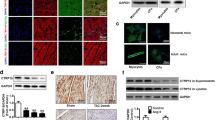
fCTRP9 is cleaved by MMP9, and overexpression of MMP9 enhances the cardioprotective effects of fCTRP9 against MI
Studies were conducted to investigate whether cleavage of fCTRP9 occurs in serum and its involvement in cardioprotection. Incubation with mice serum resulted in the cleavage of fCTRP9 into approximately 25 kDa and 15 kDa fragments (Fig. 3A). The molecular weights of these two bands corresponded to those of nCTRP9 and gCTRP9. To determine if MMP9 plays a role in the cleavage of fCTRP9, we treated fCTRP9 protein with MMP9 in vitro. As expected, fCTRP9 was cleaved by MMP9 into two fragments that closely matched gCTRP9 and nCTRP9 based on their molecular weights (Fig. 3B). Pre-incubation with MMP9 negated the promoting effect of fCTRP9 on iCFs activation and proliferation (Fig. 3C-F). To assess whether overexpression of MMP9 enhances the protective effect of fCTRP9 against MI, we induced overexpression of MMP9 in mice heart (Fig. 3G). This overexpression significantly enhanced the cleavage of fCTRP9 observed in mouse serum (Fig. 3H). Furthermore, MMP9 overexpression further improved cardiac systolic function in MI mice with increased EF compared with that in the NC + fCTRP9 group (Fig. 3I). Additionally, overexpressing MMP9 reduced both infarct size and lung-to-body weight ratio (Fig. 3J-K). Moreover, treatment with fCTRP9 in MMP9 overexpression mice led to a reduction in cardiac hypertrophy, evidenced by decreases in both heart weight relative to tibial length ratio and mean CSA measurements (Fig. 3L-M). Overexpression of MMP9 further decreased instances of cardiac fibrosis as well (Fig. 3N). These findings suggest that fCTRP9 is cleaved by MMP9; additionally, its overexpression augments cardioprotective effects of fCTRP9 against MI.
fCTRP9 is cleaved by MMP9, and overexpression of MMP9 enhances the cardioprotective effects of fCTRP9 against MI. (A) Incubation with mice serum resulted cleavage of fCTRP9. (B) Incubation with MMP9 resulted cleavage of fCTRP9. C-F. Co-incubated with MMP9 increases the anti-fibrotic effect of fCTRP9 on immortalized cardiac fibroblasts (iCF) by decreasing transcription of fibrosis-related genes (D), n = 4, decreasing expression of α-smooth muscle actin (α-SMA) (E), n = 6, and reducing cell proliferation (F), n = 6. G. Overexpression of cardiac-specific MMP9 mice was established by administrating adeno-associated virus (type 9) carrying the MMP9 coding gene via tail vein injection. fCTRP9 was administered after MI surgery for 4 weeks. H. MMP9 overexpression enhanced the cleavage of fCTRP9 in mice serum. I. Cardiac ejection fraction (EF) of mice post MI surgery. n = 6. J. Lung weight (LW) to body weight (BW) ratio. n = 6. K. Infarct size of mice heart tissue. n = 6. L. Staining with wheat germ agglutinin was done to determine the mean cross-sectional area of cardiomyocytes. n = 6. M. Heart weight (HW) to tibial length (TL) ratio. n = 6. N. Cardiac fibrosis in Sham and MI mice heart detected by Masson staining of border region and distant region. n = 6. * p < 0.05, ** p < 0.01
nCTRP9 aggravates cardiac fibrosis in MI mice
To investigate the role of nCTRP9 in cardiac fibrosis, we treated Tgf-β stimulated iCFs with nCTRP9 for subsequent analysis of activation and proliferation. Our findings indicated that nCTRP9 significantly enhanced the transcription levels of Acta3, Postn, Col1a3, and increased α-SMA expression in iCFs (Fig. 4A-B). Furthermore, nCTRP9 promoted both activation and proliferation of iCFs induced by Tgf-β (Fig. 4C-D).To assess whether nCTRP9 exacerbated cardiac injury, MI mice were administered intraperitoneal injections of nCTRP9 post-surgery (Fig. 4E). The results demonstrated a decline in cardiac function following nCTRP9 treatment (Fig. 4F-G), while there was no significant change observed in infarct size after administration of nCTRP9 (Fig. 4H). Additionally, treatment with nCTRP9 aggravated MI-induced cardiac hypertrophy as evidenced by an increase in heart weight-to-tibial length ratio and mean CSA measurements (Fig. 4I-K). Moreover, administration of nCTRP9 resulted in heightened levels of cardiac fibrosis (Fig. 4L). Notably, the application of nCTRP9 did not alter serum levels of gCTRP9 or fCTRP9 in mice (Fig. 4M). These results collectively suggest that nCTRP9 plays a detrimental role by aggravating cardiac fibrosis in mice subjected to MI.
nCTRP9 aggravates cardiac fibrosis in MI mice. (A) Impact of nCTRP9 on stimulation of immortalized cardiac fibroblasts (iCF) was tested in vitro. (B) nCTRP9 up-regulates the transcription of fibrosis-related genes in iCF. n = 4. (C) nCTRP9 increases α-smooth muscle actin (α-SMA) expression of iCF. n = 6. (D) nCTRP9 increases cell proliferation of iCF. n = 6. (E) nCTRP9 was administered via intraperitoneal injection after MI surgery for 4 weeks. F-G. Cardiac ejection fraction (EF) of mice post MI surgery. n = 6. H. Infarct size of mice heart was measured post MI surgery for 4 weeks. n = 6. I. Heart weight (HW) to tibial length (TL) ratio. n = 6. J-K. Staining with wheat germ agglutinin was done to determine the mean cross-sectional area of cardiomyocytes. n = 6. L. Cardiac fibrosis in Sham and MI mice heart detected by Masson staining of border regionand distant region. n = 6. M. Serum level of gCTRP9, nCTRP9 and fCTRP9 was measured by western blotting. * p < 0.05, ** p < 0.01
nCTRP9 promotes cardiac fibrosis through the upregulation of Rap1
To investigate the key signaling pathways involved in nCTRP9-mediated exacerbation of cardiac fibrosis, RNA-seq was performed on iCFs treated with nCTRP9. The results indicated that the Rap1-AKT/MAPK/ERK pathway may modulate the transcriptional profile of iCFs (Fig. 5A-B). To confirm the critical role of Rap1 in nCTRP9-induced CFs, iCFs were treated with both nCTRP9 and ESI-05, a specific inhibitor of Rap1. Notably, nCTRP9 failed to activate or induce proliferation in iCFs when exposed to the Rap1 inhibitor (Fig. 5C). Furthermore, phosphorylation levels of ERK 1/2 and MEK 1/2 were significantly elevated in both nCTRP9-treated iCFs and mice supplemented with nCTRP9 (Fig. 5C-D). To further elucidate whether Rap1 is involved in mediating the effects of nCTRP9 on cardiac fibrosis, we employed siRNA to knock down Rap1 expression in iCFs (Fig. 5F). In this context, treatment with nCTRP9 did not lead to an increase in transcription levels of Ccn2, Postn, Acta2, or Col1a1 within the Rap1 knockdown iCFs (Fig. 5G). Additionally, knocking down Rap1 abolished the ability of nCTRP9 to enhance proliferation and activation responses in iCFs (Fig. 5H-I). Collectively, these findings suggest that nCTRP9 promotes myocardial fibrosis by upregulating expression levels of Rap1.
nCTRP9 promotes cardiac fibrosis through upregulation of Rap1. A. Heatmap of the different genes after nCTRP9 treated immortalized cardiac fibroblasts (iCF) activated with Tgf-β. B KEGG pathway enrichment of differentially expressed genes. C. ESI-05, an inhibitor of Rap1, down-regulated the transcription of fibrosis-related genes in iCF. n = 4. D. nCTRP9 increased MEK 1/2 and ERK 1/2 phosphorylation in fibroblasts. n = 4. E. nCTRP9 increased MEK 1/2 and ERK 1/2 phosphorylation in mice heart post MI surgery. n = 4. F. Rap1 knockdown efficiency in iCF. n = 4. G. Down-regulation of Rap1 attenuated the promoting effect of nCTRP9 on the transcription of fibrosis-related genes in iCF. n = 4. H. Rap1 knockdown attenuated α-smooth muscle actin (α-SMA) expression increased by nCTRP9 in iCF. n = 6. I. Rap1 knockdown attenuated the promoting effect of nCTRP9 on cell proliferation of iCF. n = 6. * p < 0.05, ** p < 0.01
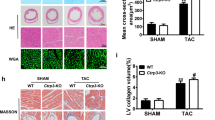
Knockdown of RAP1 abolishes cardiac fibrosis induced by nCTRP9
To investigate the role of RAP1 in the exacerbation of cardiac fibrosis induced by nCTRP9 in MI mice, we employed AAV9 to achieve a specific knockdown of Rap1 in CFs. The fibroblast-specific knockdown of Rap1 resulted in improved cardiac function, as evidenced by an increase in EF (Fig. 6A-B). However, there was no significant change observed in infarct size (Fig. 6C). Furthermore, nCTRP9 did not exacerbate cardiac hypertrophy following MI surgery in Rap1 knockdown mice, which was demonstrated by an increase in the ratio of cardiac weight to tibial length and mean CSA measurements (Fig. 6D-E). Additionally, the specific knockdown of Rap1 effectively reversed the fibrotic effects induced by nCTRP9 on cardiac tissue (Fig. 6F-G). Notably, phosphorylation levels of ERK 1/2 and MEK 1/2 were also reduced in Rap1 knockdown mice compared to controls (Fig. 6H). These findings suggest that knocking down RAP1 abolishes nCTRP9-induced cardiac fibrosis.
RAP1 knockdown abolishes cardiac fibrosis induced by nCTRP9. (A) Fibroblast-specific knockdown of Rap1 mice was established by administrating adeno-associated virus (type 9) carrying the shRNA of Rap1 via tail vein injection. nCTRP9 was administered after MI surgery for 4 weeks. (B) Cardiac ejection fraction (EF) of mice post MI surgery. n = 6. (C) Infarct size of MI mice heart was detected after surgery for 4 weeks. n = 6. (D) Heart weight (HW) to tibial length (TL) ratio. n = 6. (E) Staining with wheat germ agglutinin was done to determine the mean cross-sectional area of cardiomyocytes. n = 6. F-G. Cardiac fibrosis in Sham and MI mice heart detected by Masson staining of border region and distant region. n = 6. H. Rap1 levels in MI mice heart. n = 4. I. Knockdown of Rap1 attenuated phosphorylation increase by nCTRP9 in MI mice heart. n = 4. * p < 0.05, ** p < 0.01
Discussion
CTRP9 is a secreted protein made up of a short variable domain, followed by a collagen-like domain and a distinctive C-terminal trimerizing gC1q domain [16], primarily expressed in the adipose tissue. Research has shown that CTRP9 acts directly on heart and blood vessel cells to improve the cardiovascular system [17,18,19]. In our preliminary experiments, we reported that CTRP9 promoted endothelial cell function and improved endothelium-dependent vasorelaxation [20]. In individuals with diabetes, CTRP9 enhances microvascular endothelial function and mitigates hypoxia/reoxygenation-induced human placental vascular endothelial cell damage and mitochondrial dysfunction [21, 22]. It further decreases atherosclerosis by preventing hyperglycemia-induced endothelial cell senescence [23]. It enhances ischemia and endothelial cell function-induced revascularization [24], which causes vascular endothelial cells to respond vasoprotectively and produce angiogenesis [25]. The administration of CTRP9 by adenoviral expression systems reduces the neointimal hyperplasia in both normal and diet-induced obese mice in response to vascular damage. Furthermore, the introduction of recombinant CTRP9 protein inhibits the proliferation and chemotaxis of vascular smooth muscle cells in vitro when stimulated by mitogen stimulation [26]. CTRP9 inhibits inflammatory reactions and protects against acute cardiac damage caused by pathogenic stimuli [27]. Moreover, it increases macrophage polarization and enhances cardiac function following myocardial infarction [28]. Consistent with our previous research, CTRP9 attenuates adverse cardiac remodeling after MI [4]. Globular CTRPs are soluble CTRP variants that originate from proteolysis and only include the C1q domain. Studies have shown that cardiac tissue lysates cleave fCTRP9 into gCTRP9, which activates essential cardiac survival kinases [2, 5], and by increasing autophagic flux, globular CTRP9 shields cardiomyocytes from oxidative damage brought on by palmitic acid [29]. In our study, we found that gCTRP9 and fCTRP9 exerted different effects in the process of MI, and both gCTRP9 and fCTRP9 increased cell viability, decreased apoptosis, reduced infarct size, cardiac hypertrophy, and cardiac fibrosis, whereas gCTRP9 exerted a better protective effect than fCTRP9 against MI.
The protein family known as CTRPs, a highly conserved family of adiponectin paralogs, has four unique domains: a short variable domain, a collagen-like domain, a C-terminal C1q-like globular domain, and a signal peptide. Their structure is similar to that of adiponectin [30, 31]. Adiponectin is able to cleaved by neutrophil elastase at 5 distinct sites, 2 of which will generate adiponectin with the size corresponding to its globular head [32]. Full-length CTRP12, another analog of CTRP9, is able to cleaved by PCSK3/furinat in amino acid position Lys91 generating the gCTRP12 isoform [33]. A previous study showed that recombinant fCTRP9 was cleaved when incubated with cardiac tissue extracts, generating gCTRP9, a process inhibited by protease inhibitor cocktail [5]. In our study, we verified that fCTRP9 is cleaved by MMP9 into gCTRP9 and nCTRP9, which may partly explain why gCTRP9 exerts anti-fibrotic effects, whereas fCTRP9 promotes fibrosis.
As the primary constituents of all extracellular matrix (ECM) forms, fibrillar and non-fibrillar collagens activate discoidin domain receptors (DDRs), also known as C1q receptors. This process helps to regulate the connections between collagen and cells [34]. Mice lacking in DDR2 show reduced cutaneous wound healing, which results from decreased fibroblast proliferation and bone development due to lowered chondrocyte proliferation [35, 36]. ECM remodeling plays a crucial role in the process of re-epithelialization and is closely linked to the synthesis of MMPs. In acute human wounds, fibroblasts produce MMP2, whereas MMP9 is expressed in various wounded epithelial tissues to facilitate the healing of wounds [36]. MMP-enabled enzymatic digestion of C1q-CLR did not cause the collagen-like region’s secondary structure to disappear [37]. This indicates that the collagen-like domain may be involved in the activation and proliferation of fibroblasts, thereby promoting fibrosis. nCTRP9, collagen-like domain of fCTRP9 can promote fibrosis, thus attenuating the protective effect of fCTRP9. gCTRP9 and nCTRP9 was generated from fCTRP9, and they’re equimolar in theory. Under normal physiological conditions, supplementing of nCTRP9 had no effect on cardiac function (Fig. 4). Also, CTRP9 knockout, CTRP9 cardiac over-expression had no effect on cardiac function [7, 13]. So, we hypothesize that, under normal physiological conditions, the level of nCTRP9 and gCTRP9 does not have a significant effect on the heart.
The process of CFs proliferating and excessive extracellular matrix deposition is known as myocardial fibrosis. One of the primary processes involved in the healing process after MI is the ECM, which comprises components such as collagen I and III, fibronectin, laminin, and matrix metalloproteinases. These components are primarily generated by CFs and might manifest as reactive or replacement fibrosis [38, 39]. The process of replacement fibrosis involves the substitution of cardiomyocytes in necrotic regions by fibrotic scars, thereby averting unfavorable remodeling and ventricular wall rupture. Reactive fibrosis causes interstitial fibrosis in the infarct border zone and adjacent myocardium due to insufficient resolution of inflammation or over-repairing mechanisms [40]. The ventricular architecture and systodiastolic performance may be impacted by replacement or reactive fibrosis, which provides the pathophysiological foundation for ischemic heart failure [41, 42]. Excessive or inadequate reparative dysregulation can result in ventricular dysfunction and potentially fatal arrhythmias, which have a substantial impact on clinical results [43, 44]. Excessive cardiac fibrosis has been associated with morbidity and mortality by increasing myocardial stiffness and structural remodeling, which eventually results in heart dysfunction [45, 46]. The adverse remodeling after MI can be ameliorated by enhancing ECM degradation, reducing CFs activators or directly inhibiting CFs activation. Chymase inhibition has been shown to attenuate cardiac fibrosis by modulating MMP-9, inflammatory markers, and the eNOS pathway, as well as reducing fibrosis induced by activated chymase following MI [47]. AC261066 mitigates post-MI cardiac fibrosis through a sequential activation of RARb2 in cardiomyocytes and fibroblasts, subsequently regulating a set of genes that are responsible for decreasing oxidative stress and reactive oxygen species production [48]. Nearly 20 types of miRNAs have been identified to regulate gene expression via translational repression and/or post-transcriptional degradation, playing significant roles in inflammation and fibrosis after MI [49, 50]. Several long non-coding RNAs (lncRNAs), such as Chrf, H19, Meg3, and Wisper, participate in cardiac fibrosis by acting on either cardiomyocytes or CFs [51, 52]. Ciclopirox alleviates cardiac fibrosis post-MI by inhibiting CFs proliferation, migration, and the trans-differentiation from CFs to myofibroblasts [53]. Tartrate-resistant acid phosphatase 5 influences the proliferation, migration, and phenotypic transition of CFs; this modulation contributed to the development of myocardial fibrosis after MI through the ERK/GSK3β/β-catenin signaling pathway [54]. LIM kinase 2 promotes myofibroblast proliferation and adverse cardiac remodeling post-MI by mediating interactions between canonical and non-canonical Wnt pathways during this process [55]. CTRP9 reduces cardiac fibrosis after MI through various mechanisms. Notably, gCTRP9 mitigates adverse remodeling by enhancing survival pathways within cardiomyocytes in ischemic hearts [4]. In addition, in post-MI rats, CTRP9 reduces arrhythmogenic substrates in the atrium by partially reversing inflammation, particularly the invasion of macrophages and fibrosis [56]. In this study, we demonstrated that gCTRP9 significantly mitigates adverse remodeling by enhancing the survival of ischemic cardiomyocytes and inhibiting the proliferation of CFs. Cell experiments indicated that fCTRP9 also promotes the survival of ischemic cardiomyocytes. This finding suggests that the beneficial effects of fCTRP9 in alleviating adverse remodeling may primarily stem from its role in supporting cardiomyocyte survival. In contrast, nCTRP does not influence cardiomyocyte apoptosis; rather, it promotes proliferation of CFs, thereby exacerbating cardiac function.
Repressor activator protein 1 (Rap1) is a ubiquitous protein that exists in two isoforms, Rap1a and Rap1b. It is a member of the Ras superfamily with endogenous GTP-hydrolyzing activity [57, 58]. Rap1a and Rap1b are critical for regulating several metabolic processes. Increased and abnormal activation of Rap1 can result in tumor formation and the development of malignancy [59]. Rap1 is a small GTPase that has been shown to regulate cell proliferation, adhesion, and junction formation. This suggests that Rap1 plays an integral role in the cardiovascular signaling pathways required for cardiac function and development [57]. A novel regulator of important cardiovascular system functions, such as blood vessel development and permeability, aggregation of platelets, and cardiomyocyte survival and growth, has been identified as Rap1 [57]. According to reports, Rap1 plays a crucial role in maintaining normal vascular tone by crucially regulating eNOS, NO release, and endothelial function [60]. Deletion of Rap1 protects against myocardial ischemia/ reperfusion injury by suppressing cell apoptosis [61]. Moreover, Rap1 has been linked to the control of the extracellular matrix, as well as the migration and proliferation of particular fibroblasts [62]. Recent research has shown that renal tissue macrophage infiltration is facilitated by PP2Acα activation in macrophages through Rap1-regulated migration [62]. Rap1 is also essential for a number of integrin-mediated biological activities, including cell attachment to extracellular proteins such as collagen, laminin, fibronectin, and fibrinogen [63]. Additional research revealed that adipocyte-derived exosomes transport miR-23a-3p into cardiac fibroblasts, convert fibroblasts into myofibroblasts, and promote excessive collagen deposition by targeting RAP1. These mechanisms explain the pathological communication between dysfunctional adipose tissue and the heart [64]. In fibrotic disorders, PI3K/Akt functions as a transduction molecule that aids in the activation of myofibroblasts as well as collagen synthesis [65]. According to recent studies, phosphorylation of Akt is elevated in liver fibrosis, and this is thought to be a signal for the development of eNOS, which in turn produces NO [66]. It has also been demonstrated that the proteasomal degradation of G protein-coupled receptor kinase 2 (GRK2) can be interrupted by ERK phosphorylation on Ser-670 [67]. Increasing the levels of p-GRK2, which included by p-ERK1/2, has a negative effect by promoting the expansion of MI and fibrotic regions, as well as stimulating collagen production. Blocking ERK1/2 signaling in mouse CFs resulted in a reduction in p-GRK2 expression. These findings suggest that the PI3K/Akt and ERK signaling pathways play a role in the development of cardiac fibrosis. In our study, we found that gCTRP9 exerts anti-fibrotic effects, whereas nCTRP9 promotes myocardial fibrosis. NCTRP9 failed to increase the transcription of Ccn2, Postn, Acta2, and Col1a1 in Rap1 knockdown iCFs, and the knockdown of Rap1 abolished the ability to increase the proliferation and activation of iCFs by nCTRP9. These results suggest that nCTRP9 promotes myocardial fibrosis by upregulating Rap1 via the MEK/ERK signaling pathway, resulting in extracellular matrix (ECM) remodeling and impaired cardiac function.
Several limitations are present in this study. The precise cleavage site of MMP9 on fCTRP9 remains unclear. We attempted to isolate the MMP9 cleavage product from SDS-PAGE and perform amino acid sequencing to identify the specific cleavage site. However, due to issues related to the quantity and purity of the product, we could only infer that the cleavage site may lie between GIRGWKGDRGEKGKIGE and TLVLPKSAFTVGLTVLS (data not shown). We did not validate this finding through amino acid synonymous mutations. Additionally, it is still unknown whether there are other potential cleavage sites, further work is necessary. Furthermore, in our search for proteolytic enzymes capable of cleaving fCTRP9, we focused solely on MMP9 and furin (data not shown). Fortunately, MMP9 was able to cleave fCTRP9; however, it remains uncertain whether other proteases can also cleave fCTRP9 in vivo, necessitating further investigation. Finally, off-target effects may occur when Rap1 is knockdown using shRNA. To confirm the findings of this study, CRISPR/Cas9-specific knockout mice will be employed.
Conclusions
Our findings indicate that various forms of CTRP9 are present in the serum of MI mice, each exhibiting varying roles in post-MI cardiac protection. gCTRP9 significantly enhances cardiac function, reduces infarct size, and has anti-fibrotic properties. While fCTRP9 shares some functions with gCTRP9, its effects are weaker. In contrast, nCTRP9 does not affect infarct size and is associated with worsening cardiac function and increased fibrosis. These results highlight the critical role of CTRP9 in regulating cardiac function and suggest potential therapeutic strategies for MI treatment.
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information files.
Abbreviations
- CF:
-
Cardiac fibroblast
- CSA:
-
Mean cross-sectional area
- CTRP9:
-
C1q/tumor necrosis factor-related protein 9
- EF:
-
Ejection fraction
- gCTRP9:
-
Globular domain of CTRP9
- gC1q:
-
Globular C1q
- HW/TL:
-
Heart weight/tibial length
- iCF:
-
Immortalized cardiac fibroblast
- LVEDD:
-
Left ventricular end-diastolic diameter
- LW/BW:
-
Lung weight/body weight
- MI:
-
Myocardial infarction
- MMP9:
-
Matrix metalloproteinase-9
- nCTRP9:
-
N-terminal domain of CTRP9
- NRCM:
-
Neonatal rat cardiomyocyte
- Rap1:
-
Repressor activator protein 1
- SI:
-
Simulated ischemic
References
Salari N, Morddarvanjoghi F, Abdolmaleki A, Rasoulpoor S, Khaleghi AA, Hezarkhani LA, et al. The global prevalence of myocardial infarction: a systematic review and meta-analysis. BMC Cardiovasc Disord. 2023;23(1):206. https://doi.org/10.1186/s12872-023-03231-w.
Liu D, Gu G, Gan L, Yan W, Zhang Z, Yao P, et al. Identification of a CTRP9 C-Terminal polypeptide capable of enhancing bone-derived mesenchymal stem cell cardioprotection through promoting angiogenic exosome production. Redox Biol. 2021;41. https://doi.org/10.1016/j.redox.2021.101929.
Nakamura K, Fuster JJ, Walsh K, Adipokines. A link between obesity and cardiovascular disease. J Cardiol. 2014;63(4):250–9. https://doi.org/10.1016/j.jjcc.2013.11.006.
Sun Y, Yi W, Yuan Y, Lau WB, Yi D, Wang X, et al. C1q/tumor necrosis factor–related protein-9, a novel adipocyte-derived cytokine, attenuates adverse remodeling in the ischemic mouse heart via protein kinase a activation. Circulation. 2013;128(11suppl1):13–20. https://doi.org/10.1161/circulationaha.112.000010.
Yuan Y, Lau WB, Su H, Sun Y, Yi W, Du Y, et al. C1q-TNF-related protein-9, a novel cardioprotetcive cardiokine, requires proteolytic cleavage to generate a biologically active globular domain isoform. Am J Physiology-Endocrinology Metabolism. 2015;308(10):E891–8. https://doi.org/10.1152/ajpendo.00450.2014.
Gao E, Lei YH, Shang X, Huang ZM, Zuo L, Boucher M, et al. A novel and efficient model of coronary artery ligation and myocardial infarction in the mouse. Circul Res. 2010;107(12):1445–53. https://doi.org/10.1161/circresaha.110.223925.
Zhao D, Feng P, Sun Y, Qin Z, Zhang Z, Tan Y, et al. Cardiac-derived CTRP9 protects against myocardial ischemia/reperfusion injury via calreticulin-dependent Inhibition of apoptosis. Cell Death Dis. 2018;9(7):723. https://doi.org/10.1038/s41419-018-0726-3.
Zhang B, Shi L, Tan Y, Zhou Y, Cui J, Song Y, et al. Forkhead box O6 (FoxO6) promotes cardiac pathological remodeling and dysfunction by activating Kif15–TGF-β1 under aggravated afterload. MedComm. 2023;4(5):e383. https://doi.org/10.1002/mco2.383.
Zhang B, Zhang P, Tan Y, Feng P, Zhang Z, Liang H, et al. C1q-TNF-related protein-3 attenuates pressure overload-induced cardiac hypertrophy by suppressing the p38/CREB pathway and p38-induced ER stress. Cell Death Dis. 2019;10(7):520. https://doi.org/10.1038/s41419-019-1749-0.
Jabłońska-Trypuć A, Matejczyk M, Rosochacki S. Matrix metalloproteinases (MMPs), the main extracellular matrix (ECM) enzymes in collagen degradation, as a target for anticancer drugs. J Enzyme Inhib Med Chem. 2016;31(sup1):177–83. https://doi.org/10.3109/14756366.2016.1161620.
Tan Y, Li M, Wu G, Lou J, Feng M, Xu J, et al. Short-term but not long-term high fat diet feeding protects against pressure overload-induced heart failure through activation of mitophagy. Life Sci. 2021;272:119242. https://doi.org/10.1016/j.lfs.2021.119242.
Yokota T, McCourt J, Ma F, Ren S, Li S, Kim T-H, et al. Type V collagen in Scar tissue regulates the size of Scar after heart injury. Cell. 2020;182(3):545–62. https://doi.org/10.1016/j.cell.2020.06.030.
Tan Y, Feng P, Feng L, Shi L, Song Y, Yang J, et al. Low-dose exercise protects the heart against established myocardial infarction via IGF‐1‐upregulated CTRP9 in male mice. MedComm. 2023;4(6):e411. https://doi.org/10.1002/mco2.411.
Zhang B, Tan Y, Zhang Z, Feng P, Ding W, Wang Q, et al. Novel PGC-1α/ATF5 Axis partly activates UPRmt and mediates cardioprotective role of Tetrahydrocurcumin in pathological cardiac hypertrophy. Oxidative Med Cell Longev. 2020;2020:1–21. https://doi.org/10.1155/2020/9187065.
Zhang M, Tan Y, Song Y, zhu M, Zhang B, Chen C, et al. GLUT4 mediates the protective function of Gastrodin against pressure overload-induced cardiac hypertrophy. Biomed Pharmacother. 2023;161:114324. https://doi.org/10.1016/j.biopha.2023.114324.
Schanbacher C, Hermanns HM, Lorenz K, Wajant H, Lang I. Complement 1q/tumor necrosis factor-related proteins (CTRPs): structure, receptors and signaling. Biomedicines. 2023;11(2):559. https://doi.org/10.3390/biomedicines11020559.
Yang Y, Li Y, Ma Z, Jiang S, Fan C, Hu W, et al. A brief glimpse at CTRP3 and CTRP9 in lipid metabolism and cardiovascular protection. Prog Lipid Res. 2016;64:170–7. https://doi.org/10.1016/j.plipres.2016.10.001.
Lin JM, Hsu CH, Chen JC, Kao SH, Lin YC. BCL-6 promotes the methylation of miR‐34a by recruiting EZH2 and upregulating CTRP9 to protect ischemic myocardial injury. BioFactors. 2021;47(3):386–402. https://doi.org/10.1002/biof.1704.
Si Y, Fan W, Sun L. A review of the relationship between CTRP family and coronary artery disease. Curr Atheroscler Rep. 2020;22(6):22. https://doi.org/10.1007/s11883-020-00840-0.
Zheng Q, Yuan Y, Yi W, Lau WB, Wang Y, Wang X, et al. C1q/Ttnf-related proteins, a family of novel adipokines, induce vascular relaxation through the adiponectin receptor-1/AMPK/eNOS/Nitric oxide signaling pathway. Arterioscler Thromb Vasc Biol. 2011;31(11):2616–23. https://doi.org/10.1161/atvbaha.111.231050.
Zhu L, Chen S, Dai X. CTRP9 alleviates hypoxia/reoxygenation-induced human placental vascular endothelial cells impairment and mitochondrial dysfunction through activating AMPK/Nrf2 signaling. Tissue Cell. 2023;85:102217. https://doi.org/10.1016/j.tice.2023.102217.
Yan Z, Cao X, Wang C, Liu S, Li Y, Lu G, et al. C1q/tumor necrosis factor-related protein-3 improves microvascular endothelial function in diabetes through the AMPK/eNOS/NO· signaling pathway. Biochem Pharmacol. 2022;195:114745. https://doi.org/10.1016/j.bcp.2021.114745.
Wang G, Han B, Zhang R, Liu Q, Wang X, Huang X, et al. C1q/tnf-related protein 9 attenuates atherosclerosis by inhibiting hyperglycemia-induced endothelial cell senescence through the AMPKα/KLF4 signaling pathway. Front Pharmacol. 2021;12:758792. https://doi.org/10.3389/fphar.2021.758792.
Yamaguchi S, Shibata R, Ohashi K, Enomoto T, Ogawa H, Otaka N, et al. C1q/tnf-related protein 9 promotes revascularization in response to ischemia via an eNOS-dependent manner. Front Pharmacol. 2020;11:1313. https://doi.org/10.3389/fphar.2020.01313.
Lee SM, Lee JW, Kim I, Woo D-C, Pack C-G, Sung YH, et al. Angiogenic adipokine C1q-TNF–related protein 9 ameliorates myocardial infarction via histone deacetylase 7–mediated MEF2 activation. Sci Adv. 2022;8(48):eabq0898. https://doi.org/10.1126/sciadv.abq0898.
Uemura Y, Shibata R, Ohashi K, Enomoto T, Kambara T, Yamamoto T, et al. Adipose-derived factor CTRP9 attenuates vascular smooth muscle cell proliferation and neointimal formation. FASEB J. 2012;27(1):25–33. https://doi.org/10.1096/fj.12-213744.
Kambara T, Shibata R, Ohashi K, Matsuo K, Hiramatsu-Ito M, Enomoto T, et al. C1q/tumor necrosis factor-related protein 9 protects against acute myocardial injury through an adiponectin receptor I-AMPK-dependent mechanism. Mol Cell Biol. 2023;35(12):2173–85. https://doi.org/10.1128/mcb.01518-14.
Liu M, Yin L, Li W, Hu J, Wang H, Ye B, et al. C1q/TNF-related protein‐9 promotes macrophage polarization and improves cardiac dysfunction after myocardial infarction. J Cell Physiol. 2019;234(10):18731–47. https://doi.org/10.1002/jcp.28513.
Zuo A, Li J, Zhao X, Li T, Lei S, Chen J, et al. Globular CTRP9 protects cardiomyocytes from palmitic acid-induced oxidative stress by enhancing autophagic flux. Chemico-Biol Interact. 2020;329:109094. https://doi.org/10.1016/j.cbi.2020.109094.
Wong GW, Wang J, Hug C, Tsao T-S, Lodish HF. A family of Acrp30/adiponectin structural and functional paralogs. Proceedings of the National Academy of Sciences. 2004;101(28):10302–10307. https://doi.org/10.1073/pnas.0403760101
Wei Z, Peterson JM, Wong GW. Metabolic regulation by c1q/tnfF-related protein-13 (CTRP13). J Biol Chem. 2011;286(18):15652–65. https://doi.org/10.1074/jbc.M110.201087.
Waki H, Yamauchi T, Kamon J, Kita S, Ito Y, Hada Y, et al. Generation of globular fragment of adiponectin by leukocyte elastase secreted by monocytic cell line THP-1. Endocrinology. 2005;146(2):790–6. https://doi.org/10.1210/en.2004-1096.
Wei Z, Lei X, Seldin MM, Wong GW. Endopeptidase cleavage generates a functionally distinct isoform of C1q/Tumor necrosis Factor-related Protein-12 (CTRP12) with an altered oligomeric state and signaling specificity. J Biol Chem. 2012;287(43):35804–14. https://doi.org/10.1074/jbc.M112.365965.
Shrivastava A, Radziejewski C, Campbell E, Kovac L, McGlynn M, Ryan TE, et al. An orphan receptor tyrosine kinase family whose members serve as nonintegrin collagen receptors. Mol Cell. 1997;1(1):25–34. https://doi.org/10.1016/s1097-2765(00)80004-0.
Olaso E, Lin H-C, Wang L-H, Friedman SL. Impaired dermal wound healing in discoidin domain receptor 2-deficient mice associated with defective extracellular matrix remodeling. Fibrogenesis Tissue Repair. 2011;4(1):5. https://doi.org/10.1186/1755-1536-4-5.
Hayuningtyas RA, Han M, Choi S, Kwak MS, Park IH, Lee J-H, et al. The collagen structure of C1q induces wound healing by engaging discoidin domain receptor 2. Mol Med. 2021;27(1):125. https://doi.org/10.1186/s10020-021-00388-y.
Ruiz S, Henschen-Edman AH, Nagase H, Tenner AJ. Digestion of C1q collagen-like domain with MMPs-1, -2, -3, and– 9 further defines the sequence involved in the stimulation of neutrophil superoxide production. J Leukoc Biol. 1999;66(3):416–22. https://doi.org/10.1002/jlb.66.3.416.
Talman V, Ruskoaho H. Cardiac fibrosis in myocardial infarction—from repair and remodeling to regeneration. Cell Tissue Res. 2016;365(3):563–81. https://doi.org/10.1007/s00441-016-2431-9.
Scalise RFM, De Sarro R, Caracciolo A, Lauro R, Squadrito F, Carerj S, et al. Fibrosis after myocardial infarction: an overview on cellular processes, molecular pathways, clinical evaluation and prognostic value. Med Sci. 2021;9(1):16. https://doi.org/10.3390/medsci9010016.
van Amerongen MJ, Harmsen MC, van Rooijen N, Petersen AH, van Luyn MJA. Macrophage depletion impairs wound healing and increases left ventricular remodeling after myocardial injury in mice. Am J Pathol. 2007;170(3):818–29. https://doi.org/10.2353/ajpath.2007.060547.
Prabhu S. Post-infarction ventricular remodeling: an array of molecular events. J Mol Cell Cardiol. 2005;38(4):547–50. https://doi.org/10.1016/j.yjmcc.2005.01.014.
Sutton MGSJ, Sharpe N. Left ventricular remodeling after myocardial infarction. Circulation. 2000;101(25):2981–8. https://doi.org/10.1161/01.Cir.101.25.2981.
López B, Ravassa S, Moreno MU, José GS, Beaumont J, González A, et al. Diffuse myocardial fibrosis: mechanisms, diagnosis and therapeutic approaches. Nat Reviews Cardiol. 2021;18(7):479–98. https://doi.org/10.1038/s41569-020-00504-1.
Tikhomirov R, Reilly-O’Donnell B, Catapano F, Faggian G, Gorelik J, Martelli F, et al. Exosomes: from potential culprits to new therapeutic promise in the setting of cardiac fibrosis. Cells. 2020;9(3):592. https://doi.org/10.3390/cells9030592.
Travers JG, Kamal FA, Robbins J, Yutzey KE, Blaxall BC. Cardiac fibrosis. Circul Res. 2016;118(6):1021–40. https://doi.org/10.1161/circresaha.115.306565.
Molaei A, Molaei E, Hayes AW, Karimi G. Mas receptor: a potential strategy in the management of ischemic cardiovascular diseases. Cell Cycle. 2023;22(13):1654–74. https://doi.org/10.1080/15384101.2023.2228089.
Oyamada S, Bianchi C, Takai S, Chu LM, Sellke FW. Chymase Inhibition reduces infarction and matrix Metalloproteinase-9 activation and attenuates inflammation and fibrosis after acute myocardial ischemia/reperfusion. J Pharmacol Exp Ther. 2011;339(1):143–51. https://doi.org/10.1124/jpet.111.179697.
Tang X-H, Gambardella J, Jankauskas S, Wang X, Santulli G, Gudas LJ, et al. A retinoic acid receptor Β2 agonist improves cardiac function in a heart failure model. J Pharmacol Exp Ther. 2021;379(2):182–90. https://doi.org/10.1124/jpet.121.000806.
Varzideh F, Kansakar U, Donkor K, Wilson S, Jankauskas SS, Mone P, et al. Cardiac remodeling after myocardial infarction: functional contribution of MicroRNAs to inflammation and fibrosis. Front Cardiovasc Med. 2022;9. https://doi.org/10.3389/fcvm.2022.863238.
Wang X, Morelli MB, Matarese A, Sardu C, Santulli G. Cardiomyocyte-derived Exosomal microRNA‐92a mediates post‐ischemic myofibroblast activation both in vitro and ex vivo. ESC Heart Fail. 2020;7(1):285–9. https://doi.org/10.1002/ehf2.12584.
Ilieva M, Uchida S. Long Non-Coding RNAs in cardiac and pulmonary fibroblasts and fibrosis. Non-Coding RNA. 2022;8(4). https://doi.org/10.3390/ncrna8040053.
Kishore R, Magadum A. Cell-Specific mRNA therapeutics for cardiovascular diseases and regeneration. J Cardiovasc Dev Disease. 2024;11(2). https://doi.org/10.3390/jcdd11020038.
Subbaiah KCV, Wu J, Tang WHW, Yao P. Ciclopirox Inhibition of eIF5A hypusination attenuates fibroblast activation and cardiac fibrosis. J Cardiovasc Dev Disease. 2023;10(2). https://doi.org/10.3390/jcdd10020052.
Yang S, Pei L, Huang Z, Zhong Y, Li J, Hong Y, et al. Inhibition of tartrate-resistant acid phosphatase 5 can prevent cardiac fibrosis after myocardial infarction. Mol Med. 2024;30(1). https://doi.org/10.1186/s10020-024-00856-1.
Gong C, Chang L, Huang R, Sun X, Liu Y, Wu S, et al. LIM kinase 2 activates cardiac fibroblasts and exacerbates postinfarction left ventricular remodeling via crosstalk between the canonical and non-canonical Wnt pathways. Pharmacol Res. 2024;208. https://doi.org/10.1016/j.phrs.2024.107347.
Liu M, Li W, Wang H, Yin L, Ye B, Tang Y, et al. CTRP9 ameliorates atrial inflammation, fibrosis, and vulnerability to atrial fibrillation in post-myocardial infarction rats. J Am Heart Association. 2019;8(21):e013133. https://doi.org/10.1161/jaha.119.013133.
Jeyaraj SC, Unger NT, Chotani MA. Rap1 GTPases: an emerging role in the cardiovasculature. Life Sci. 2011;88(15–16):645–52. https://doi.org/10.1016/j.lfs.2011.01.023.
Jaśkiewicz A, Pająk B, Orzechowski A. The many faces of Rap1 GTPase. Int J Mol Sci. 2018;19(10). https://doi.org/10.3390/ijms19102848.
Itoh M, Nelson CM, Myers CA, Bissell MJ. Rap1 integratesyissue Polarity, lumen formation, and tumorigenic potential in human breast epithelial cells. Cancer Res. 2007;67(10):4759–66. https://doi.org/10.1158/0008-5472.Can-06-4246.
Lakshmikanthan S, Zheng X, Nishijima Y, Sobczak M, Szabo A, Vasquez-Vivar J, et al. Rap1 promotes endothelial mechanosensing complex formation, NO release and normal endothelial function. EMBO Rep. 2015;16(5):628–37. https://doi.org/10.15252/embr.201439846.
Cai Y, Ying F, Liu H, Ge L, Song E, Wang L, et al. Deletion of Rap1 protects against myocardial ischemia/reperfusion injury through suppressing cell apoptosis via activation of STAT3 signaling. FASEB J. 2020;34(3):4482–96. https://doi.org/10.1096/fj.201901592RR.
Li N, Shan S, Li X-Q, Chen T-T, Qi M, Zhang S-N, et al. G protein-voupled receptor kinase 2 as novel therapeutic target in fibrotic diseases. Front Immunol. 2022;12. https://doi.org/10.3389/fimmu.2021.822345.
Retta SF, Balzac F, Avolio M. Rap1: A turnabout for the crosstalk between cadherins and integrins. Eur J Cell Biol. 2006;85(3–4):283–93. https://doi.org/10.1016/j.ejcb.2005.09.007.
Su M, Li W, Yuan YUE, Liu S, Liang C, Liu HE, et al. Epididymal white adipose tissue promotes angiotensin II-induced cardiac fibrosis in an exosome-dependent manner. Translational Res. 2022;248:51–67. https://doi.org/10.1016/j.trsl.2022.05.004.
Higgins DF, Ewart LM, Masterson E, Tennant S, Grebnev G, Prunotto M et al. BMP7-induced-Pten inhibits Akt and prevents renal fibrosis. Biochimica et biophysica acta (BBA) - Molecular basis of disease. 2017;1863(12):3095–104. https://doi.org/10.1016/j.bbadis.2017.09.011
Zheng Y, Wang J, Zhao T, Wang L, Wang J. Modulation of the VEGF/AKT/eNOS signaling pathway to regulate liver angiogenesis to explore the anti-hepatic fibrosis mechanism of curcumol. J Ethnopharmacol. 2021;280:114480. https://doi.org/10.1016/j.jep.2021.114480.
Elorza A, Penela P, Sarnago S, Mayor F. MAPK-dependent degradation of G protein-coupled receptor kinase 2. J Biol Chem. 2003;278(31):29164–73. https://doi.org/10.1074/jbc.M304314200.
Funding
This work was supported by the National Natural Science Foundation of China (grant nos. 81870266, 82170336, 82270286, and 82000227), National Key R&D Program of China (2022YFC2402804), Science and Technology Research and Development Program of Shaanxi Province(2023-ZDLSF-39).
Author information
Authors and Affiliations
Contributions
YS, WY and YT designed the study. YT, GC, HL, JX, and LS conducted the experiments and collected the data. DY, YL, PL and YZ analyzed the data. YT, GC, HL, and BZ drafted the manuscript. All authors revised and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All animal experiments were performed in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals and approved by the Fourth Military Medical University Committee of Animal Care.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tan, Y., Li, H., Cao, G. et al. N-terminal domain of CTRP9 promotes cardiac fibroblast activation in myocardial infarction via Rap1/Mek/Erk pathway. J Transl Med 23, 300 (2025). https://doi.org/10.1186/s12967-025-06274-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1186/s12967-025-06274-z